
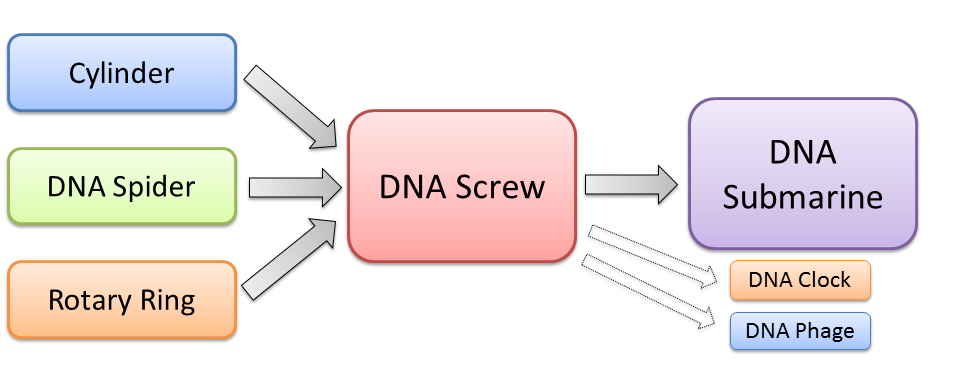
The Komaba team will develop a new molecular motor based on DNA origami. Our new motor, which we named DNA Screw, has a rotary movement using a cylinder and a ring made of DNA strands. The rotary movement is exploited from DNA walkers which advance in a certain direction by cutting a DNA footing with DNAzyme. The ring is connected to DNA walker, and on the surface of the cylinder, DNA footings are set in a spiral line. DNA Screw is more stable and easier to operate than other biomolecular motor like kinesin, because it is "artificial" products of "DNA material". Therefore this technology will greatly contribute to the practical realization of tricky movement of biomolecular-machine. We are, in particular, aiming to make a DNA structure from the idea of this screw which moves like a submarine and transports some materials. Moreover, the function of chemotaxis which microbes obtain would be created only with DNAs by controlling this DNA submarine with sensors made by DNA Origami, and this is a giant step toward creating fully autonomous DNA robots.
There were many previous studies related to the spontaneous activities of biomolecules about Kinesin, which is a class of motor proteins, and walking DNA robots. But genuine nano-scale DNA motors which rotate at the stable speed were not yet created. In our project, we are aiming at creating the DNA screw system to achieve this goal. The rotation system is used to create the complex motion with any devices, such as drills, screws and clocks. Therefore we have thought that the nano-scale rotation system enables us to extend the future of DNA engineering. The DNA screw has many strong points. DNA screw is able to embed in any other DNA structures and to be assembled into more complex structures easily, because we can take engineering approaches to make DNA structures. And, the size of the structure can be easily scaled. In addition, DNA is a stable material than protein and can be used in various environments (ex. Temperature, pH and salt-density).
The motor made in this project comprises three parts; the cylinder as an axis, the DNA Spiders as a source of power, and the rotating ring. In the first phase in the project, we design each part and confirm if each part is actually formed as we design by an Atomic Force Microscope. Then, in the second phase, we develop a method to combine those three parts, which results in construction of the DNA screw.

DNA submarine is a structure which moves in a solution and transports materials. In addition, in combination with sensors using DNA Origami technology, this DNA submarine would be able to develop into autonomous DNA robots which have the function of chemotaxis.
By using the rotary motion of this DNA screw, the motion of a phage that makes a hole on a cell wall of bacteria could be imitated artifically. And in the end, it would be possible to make an artificial phage that is entirely consisted of DNA and controlled artificially.
Rotations of DNA screw can be stabilized by raising the number of DNA spiders, and then DNA screw will become a time counting machine like a clock. It would contribute to DNA computing technology by using it as a clock of CPU.
The cylinder is put in the center of the motor as an axis supporting the rotation. DNA strands of staples and a scafold are formed into a cylindrical shape using DNA Origami technology. It was designed with cadnano software, which "simplifies and enhances the process of designing three-dimensional DNA origami nanostructures"(Figure 1). In order to bind footing DNAs on its surface spirally, 10mer long DNA strands, called probes, are jutting out from the cylinder's surface. After the cylinder with the probes is formed, anchor of DNA spiders is hybridized to a specific strand at the start point on the cylinder's surface. Then the footings get connected to the probes. The footing tracks, each of which consists of three lanes of the footings, enable DNA Spiders to orbit the cylinder. In detail, there are two tracks of DNA footings and two spiders walk on each track. In addition, in order to make it easy to observe the direction of spiders' walk, a small sub-cylinder is attached at the end of the footing tracks(Figure 2). The diameter of the cylinder is 25.4 nm and height is 43.5 nm. This is calculated considering that the cylinder can be observed with an Atomic Force Microscope and that the interval between the two tracks are wide enough for spiders not to jump to next footing track (Figure 3).
Our DNA screw rotates by using DNA spiders, which is being created based on the work of Lund, et al. DNA spider is connected to the DNA ring by a “head” strand, and also DNA spider's legs are connected to strands on the surface of the DNA cylinder. DNA spider consists of a body, a head, an anchor, and three legs, all of which are going to be made by DNA. These legs advance by cutting strands with DNAzyme. DNAzyme is a DNA which cuts DNA strands like enzymes do. Because the spider's track is a spiral, DNA rings rotates as DNA spiders walk. The head is complementary to the strand stretching from a ring made of DNA.
In detail, legs and the footings are complementary strands. The footings are attached spirally to the surface of the DNA cylinder and function as a substrate of the DNA spider. DNA spiders normally move one way. Once DNAzyme cuts the tip of the footings, DNA spiders are disconnected to the footings. The footing is binding to the nearest track strand and the cutting process is occurring again.
We will use the same DNA spider’s sequences as the way Lund, et al. did; 5’ - TCT CTT CTC CGA GCC GGT CGA AAT AGT GAA AA – 3’. We are developing the body sequence of the DNA spider. When we decide them, we will order them as soon as possible.
It is the DNA ring that actually rotates. It has a shape of a cylinder short in the axial direction and is also composed with DNA Origami technology. We made the structure using cadnano (Figure4). Two 10mer long strands come up from the inner side of the ring and are connected to the DNA spiders. The diameter of the ring is 50.9 nm, and so it can hold the cylinder and spiders inside of the ring. The thickness of ring is 10.9 nm in consideration of Atomic Force Microscope visibility (Figure 5).
The DNA screw is realized by assembling the above four parts: the cylinder, footings, DNA Spiders, and the rotary ring (Figure 6).
Team Name : Komaba-Team
Institution : University of Tokyo
Fukuhara Yusuke - Major in Material Science and Engineering
Kondo Toyoki - College of Arts and Sciences
Miyazaki Yuki - Major in Applied Physics
Osamu Ishimura - College of Arts and Sciences
Shoko Yokokawa - College of Arts and Sciences
Shuhei Nakajma - College of Arts and Sciences
Soejima Tomohiro
Wang Shaoyu
coming soon...